Eureka! Control Files (.ecf)
To run the different Stages of Eureka!, the pipeline requires control files (.ecf) where Stage-specific parameters are defined (e.g. aperture size, path of the data, etc.).
In the following, we look at the contents of the ecf for Stages 1, 2, 3, 4, 5, and 6.
Stage 1
# Eureka! Control File for Stage 1: Detector Processing # Stage 1 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-1 suffix uncal # Control ramp fitting method ramp_fit_algorithm 'default' #Options are 'default', 'mean', or 'differenced' ramp_fit_max_cores 'none' #Options are 'none', quarter', 'half','all' # Pipeline stages skip_group_scale False skip_dq_init False skip_saturation False skip_ipc True #Skipped by default for all instruments skip_superbias False skip_refpix False skip_linearity False skip_persistence True #Skipped by default for Near-IR TSO skip_dark_current False skip_jump False skip_ramp_fitting False skip_gain_scale False #Pipeline stages parameters jump_rejection_threshold 4.0 #float, default is 4.0, CR sigma rejection threshold # Custom linearity reference file custom_linearity False linearity_file /path/to/custom/linearity/fits/file # Custom bias bias_correction None # Bias correction options: [mean, group_level, smooth, None] bias_group 1 # Group number options: [1, 2, ..., each] bias_smooth_length 201 # Window length when using 'smooth' bias correction custom_bias False superbias_file /path/to/custom/superbias/fits/file # Saturation update_sat_flags False # Wheter to update the saturation flags more aggressively expand_prev_group False # Expand saturation flags to previous group dq_sat_mode percentile # Options: [percentile, min, defined] dq_sat_percentile 50 # Percentile of the entire time series to use to define the saturation mask (50=median) dq_sat_columns [[0, 0], [0,0], [0,0], [0,0], [0,0]] #for dq_sat_mode = defined, user defined saturated columns # Background subtraction grouplevel_bg True ncpu 6 bg_y1 6 bg_y2 26 bg_deg 0 p3thresh 5 verbose True isplots_S1 3 nplots 5 hide_plots True # Mask curved traces masktrace True window_len 11 expand_mask 8 ignore_low 600 ignore_hi None # Manual reference pixel correction for NIRSpec PRISM when not subtracting BG refpix_corr False npix_top 8 npix_bot 8 # Project directory topdir /home/User/Data/JWST-Sim/NIRSpec/ # Directories relative to topdir inputdir Uncalibrated outputdir Stage1 # Diagnostics testing_S1 False ##### # "Default" ramp fitting settings default_ramp_fit_weighting default #Options are "default", "fixed", "interpolated", "flat", or "custom" default_ramp_fit_fixed_exponent 10 #Only used for "fixed" weighting default_ramp_fit_custom_snr_bounds [5,10,20,50,100] # Only used for "custom" weighting, array no spaces default_ramp_fit_custom_exponents [0.4,1,3,6,10] # Only used for "custom" weighting, array no spaces
suffix
Data file suffix (e.g. uncal).
ramp_fit_algorithm
Algorithm to use to fit a ramp to the frame-level images of uncalibrated files. Only default (i.e. the JWST pipeline) and mean can be used currently.
ramp_fit_max_cores
Fraction of processor cores to use to compute the ramp fits, options are none, quarter, half, all.
skip_*
If True, skip the named step.
Note
Note that some instruments and observing modes might skip a step either way! See the calwebb_detector1 docs for the list of steps run for each instrument/mode by the STScI’s JWST pipeline.
custom_linearity
Boolean. If True, allows user to supply a custom linearity correction file and overwrite the default file.
linearity_file
The fully qualified path to the custom linearity correction file to use if custom_linearity is True.
bias_correction
Method applied to correct the superbias using a scale factor (SF) when no bias pixels are available (i.e., with NIRSpec). Here, SF = (median of group)/(median of superbias), using a background region that is expand_mask pixels from the measured trace. The default option None applies no correction; group_level computes SF for every integration in bias_group; smooth applies a smoothing filter of length bias_smooth_length to the group_level SF values; and mean uses the mean SF over all integrations. For NIRSpec, we currently recommend using smooth with a bias_smooth_length that is ~15 minutes.
Note that this routine requires masking the trace; therefore, masktrace must be set to True.
bias_group
Integer or string. Specifies which group number should be used when applying the bias correction. For NIRSpec, we currently recommend using the first group (bias_group = 1). There is no group 0. Users can also specify each, which computes a unique bias correction for each group.
bias_smooth_length
Integer. When bias_correction = smooth, this value is used as the window length during smoothing across integrations.
custom_bias
Boolean, allows user to supply a custom superbias file and overwrite the default file.
superbias_file
The fully qualified path to the custom superbias file to use if custom_bias is True.
update_sat_flags
Boolean, allows user to have more control over saturation flags. Must be True to use the settings expand_prev_group, dq_sat_mode, and dq_sat_percentile or dq_sat_columns.
expand_prev_group
Boolean, if a given group is saturated, this option will mark the previous group as saturated as well.
dq_sat_mode
Method to use for updating the saturation flags. Options are percentile (a pixel must be saturated in this percent of integrations to be marked as saturated), min, and defined (user can define which columns are saturated in a given group)
dq_sat_percentile
If dq_sat_mode = percentile, percentile threshold to use
dq_sat_columns
If dq_sat_mode = defined, list of columns. Should have length Ngroups, each element containing a list of the start and end column to mark as saturated
grouplevel_bg
Boolean, runs background subtraction at the group level (GLBS) prior to ramp fitting.
ncpu
Number of cpus to use for GLBS
bg_y1
The pixel number for the end of the bottom background region. The background region goes from the bottom of the subarray to this pixel.
bg_y2
The pixel number for the start of the top background region. The background region goes from this pixel to the top of the subarray.
bg_deg
See Stage 3 inputs
p3thresh
See Stage 3 inputs
verbose
See Stage 3 inputs
isplots_S1
Sets how many plots should be saved when running Stage 1. A full description of these outputs is available here: Stage 3 Output
nplots
See Stage 3 inputs
hide_plots
See Stage 3 inputs
bg_disp
Set True to perform row-by-row background subtraction (only useful for NIRCam).
bg_x1
Left edge of exclusion region for row-by-row background subtraction.
bg_x2
Right edge of exclusion region for row-by-row background subtraction.
masktrace
Boolean, creates a mask centered on the trace prior to GLBS for curved traces
window_len
Smoothing length for the trace location
expand_mask
Aperture (in pixels) around the trace to mask
ignore_low
Columns below this index will not be used to create the mask
ignore_hi
Columns above this index will not be used to create the mask
refpix_corr
Boolean, runs a custom ROEBA (Row-by-row, Odd-Even By Amplifier) routine for PRISM observations which do not have reference pixels within the subarray.
npix_top
Number of rows to use for ROEBA routine along the top of the subarray
npix_bot
Number of rows to use for ROEBA routine along the bottom of the subarray
topdir + inputdir
The path to the directory containing the Stage 0 JWST data (uncal.fits). Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 1 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
testing_S1
If True, only a single file will be used, outputs won’t be saved, and plots won’t be made. Useful for making sure most of the code can run.
default_ramp_fit_weighting
Define the method by which individual frame pixels will be weighted during the default ramp fitting process. The is specifically for the case where ramp_fit_algorithm is default. Options are default, fixed, interpolated, flat, or custom.
default: Slope estimation using a least-squares algorithm with an “optimal” weighting, see the ramp_fitting docs.
In short this weights each pixel, \(i\), within a slope following \(w_i = (i - i_{midpoint})^P\), where the exponent \(P\) is selected depending on the estimated signal-to-noise ratio of each pixel (see link above).
fixed: As with default, except the weighting exponent \(P\) is fixed to a precise value through the default_ramp_fit_fixed_exponent entry
interpolated: As with default, except the SNR to \(P\) lookup table is converted to a smooth interpolation.
flat: As with default, except the weighting equation is no longer used, and all pixels are weighted equally.
custom: As with default, except a custom SNR to \(P\) lookup table can be defined through the default_ramp_fit_custom_snr_bounds and default_ramp_fit_custom_exponents (see example .ecf file).
Stage 2
A full description of the Stage 2 Outputs is available here: Stage 2 Output
# Eureka! Control File for Stage 2: Data Reduction # Stage 2 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-2 suffix rateints # Data file suffix # Subarray height in cross-dispersion direction # NIRSpec slit_y_low 0 # Use None for default parameter (0) slit_y_high 32 # Use None for default parameter (16) # NIRCam grism tsgrism_extract_height 128 # Use None for default parameter (64) # Modify the existing file to change the dispersion extraction - FIX: DOES NOT WORK CURRENTLY waverange_start 6e-08 # Use None to rely on the default parameters waverange_end 6e-06 # Use None to rely on the default parameters # Note: different instruments and modes will use different steps by default skip_bkg_subtract True # Not run for TSO observations skip_imprint_subtract True # Not run for NIRSpec Fixed Slit skip_msa_flagging True # Not run for NIRSpec Fixed Slit skip_extract_2d False skip_srctype False skip_master_background True # Not run for NIRSpec Fixed Slit skip_wavecorr False skip_flat_field True # ***NOTE*** At the time the NIRSpec ERS Hackathon simulated data was created, this step did not work correctly and is by default turned off. skip_straylight True # Not run for NIRSpec Fixed Slit skip_fringe True # Not run for NIRSpec Fixed Slit skip_pathloss True # Not run for TSO observations skip_barshadow True # Not run for NIRSpec Fixed Slit skip_photom True # Recommended to skip to get better uncertainties out of Stage 3. skip_resample True # Not run for TSO observations skip_cube_build True # Not run for NIRSpec Fixed Slit skip_extract_1d False # Diagnostics testing_S2 False hide_plots False # If True, plots will automatically be closed rather than popping up # Project directory topdir /home/User/Data/JWST-Sim/NIRSpec/ # Directories relative to topdir inputdir Stage1 outputdir Stage2
suffix
Data file suffix (e.g. rateints).
Note
Note that other Instruments might used different suffixes!
slit_y_low & slit_y_high
Controls the cross-dispersion extraction for NIRSpec. Use None to rely on the default parameters.
tsgrism_extract_height
Controls the cross-dispersion extraction height for NIRCam (default is 64 pixels).
waverange_start & waverange_end
Modify the existing file to change the dispersion extraction (DO NOT CHANGE).
skip_*
If True, skip the named step.
Note
Note that some instruments and observing modes might skip a step either way! See the calwebb_spec2 docs for the list of steps run for each instrument/mode by the STScI’s JWST pipeline.
testing_S2
If True, outputs won’t be saved and plots won’t be made. Useful for making sure most of the code can run.
hide_plots
If True, plots will automatically be closed rather than popping up on the screen.
topdir + inputdir
The path to the directory containing the Stage 1 JWST data. Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 2 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
Stage 3
# Eureka! Control File for Stage 3: Data Reduction
# Stage 3 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-3
ncpu 1 # Number of CPUs
nfiles 1 # The number of data files to analyze simultaneously
max_memory 0.5 # The maximum fraction of memory you want utilized by read-in frames (this will reduce nfiles if need be)
suffix calints # Data file suffix
calibrated_spectra False # Set True to generate flux-calibrated spectra/photometry in mJy
# Set False to convert to electrons
# Subarray region of interest
ywindow [2,28] # Vertical axis as seen in DS9
xwindow [60,410] # Horizontal axis as seen in DS9
src_pos_type gaussian # Determine source position when not given in header (Options: header, gaussian, weighted, max, or hst)
record_ypos True # Option to record the y position and width for each integration (only records if src_pos_type is gaussian)
dqmask True # Mask pixels with an odd entry in the DQ array
expand 1 # Super-sampling factor along cross-dispersion direction
# Background parameters
bg_hw 7 # Half-width of exclusion region for BG subtraction (relative to source position)
bg_thresh [10,10] # Double-iteration X-sigma threshold for outlier rejection along time axis
bg_deg 1 # Polynomial order for column-by-column background subtraction, -1 for median of entire frame
p3thresh 10 # X-sigma threshold for outlier rejection during background subtraction
# Spectral extraction parameters
spec_hw 6 # Half-width of aperture region for spectral extraction (relative to source position)
fittype smooth # Method for constructing spatial profile (Options: smooth, meddata, poly, gauss, wavelet, or wavelet2D)
median_thresh 5 # Sigma threshold when flagging outliers in median frame, when fittype=meddata and window_len > 1
window_len 13 # Smoothing window length, for median frame or when fittype = smooth or meddata (when computing median frame). Can set to 1 for no smoothing when computing median frame for fittype=meddata.
prof_deg 3 # Polynomial degree, when fittype = poly
p5thresh 10 # X-sigma threshold for outlier rejection while constructing spatial profile
p7thresh 60 # X-sigma threshold for outlier rejection during optimal spectral extraction
# G395H curvature treatment
curvature None # How to manage the curved trace on the detector (Options: None, correct). Use correct for NIRSpec/G395.
# Diagnostics
isplots_S3 3 # Generate few (1), some (3), or many (5) figures (Options: 1 - 5)
nplots 5 # How many of each type of figure do you want to make per file?
vmin 0.97 # Sets the vmin of the color bar for Figure 3101.
vmax 1.03 # Sets the vmax of the color bar for Figure 3101.
time_axis 'y' # Determines whether the time axis in Figure 3101 is along the y-axis ('y') or the x-axis ('x')
testing_S3 False # Boolean, set True to only use last file and generate select figures
hide_plots False # If True, plots will automatically be closed rather than popping up
save_output True # Save outputs for use in S4
verbose True # If True, more details will be printed about steps
# Project directory
topdir /home/User/Data/JWST-Sim/NIRSpec/
# Directories relative to topdir
inputdir Stage2 # The folder containing the outputs from Eureka!'s S2 or JWST's S2 pipeline (will be overwritten if calling S2 and S3 sequentially)
outputdir Stage3
ncpu
Sets the number of cores being used when Eureka! is executed.
Currently, the only parallelized part of the code is the background subtraction for every individual integration and is being initialized in s3_reduce.py with:
util.BGsubtraction
nfiles
Sets the maximum number of data files to analyze batched together.
max_memory
Sets the maximum memory fraction (0–1) that should be used by the loaded in data files. This will reduce nfiles if needed. Note that more RAM than this may be used during operations like sigma clipping, so you’re best off setting max_memory <= 0.5.
suffix
If your data directory (topdir + inputdir, see below) contains files with different data formats, you want to consider setting this variable.
E.g.: Simulated NIRCam Data:
Stage 2 - For NIRCam, Stage 2 consists of the flat field correction, WCS/wavelength solution, and photometric calibration (counts/sec -> MJy). Note that this is specifically for NIRCam: the steps in Stage 2 change a bit depending on the instrument. The Stage 2 outputs are roughly equivalent to a “flt” file from HST.
Stage 2 Outputs/*calints.fits- Fully calibrated images (MJy) for each individual integration. This is the one you want if you’re starting with Stage 2 and want to do your own spectral extraction.Stage 2 Outputs/*x1dints.fits- A FITS binary table containing 1D extracted spectra for each integration in the “calint” files.
As we want to do our own spectral extraction, we set this variable to calints.
Note
Note that other Instruments might used different suffixes!
photometry
Only used for photometry analyses. Set to True if the user wants to analyze a photometric dataset.
calibrated_spectra
An optional input parameter. If False (default), convert the units of the images to electrons for easy noise estimation. If True (useful for flux-calibrated spectroscopy/photometry), the units of the images will be converted to mJy.
poly_wavelength
If True, use an updated polynomial wavelength solution for NIRCam longwave spectroscopy instead of the linear wavelength solution currently assumed by STScI.
gain
Optional input. If None (default), automatically use reference files or FITS header to compute the gain. If not None AND gainfile is None, this specifies the gain in units of e-/ADU or e-/DN. The gain variable can either be a single value that is applied to the entire frame or an array of the same shape as the subarray you’re using.
gainfile
Optional input. If None (default), automatically use reference files or FITS header to compute the gain. If not None, this should be a fully qualified path to a FITS file with all the same formatting as the GAIN files hosted by the CRDS. This can be used to force the use of a different version of the reference file or the use of a customized reference file.
photfile
Optional input. If None (default), automatically use reference files or FITS header to compute between brightness units (e.g. MJy/sr) to ADU or DN if required. If not None, this should be a fully qualified path to a FITS file with all the same formatting as the PHOTOM files hosted by the CRDS. This can be used to force the use of a different version of the reference file or the use of a customized reference file.
hst_cal
Only used for HST analyses. The fully qualified path to the folder containing HST calibration files.
horizonsfile
Only used for HST analyses. The path with respect to hst_cal to the horizons file you’ve downloaded from https://ssd.jpl.nasa.gov/horizons/app.html#/. To get a new horizons file on that website, 1. Select “Vector Table”, 2. Select “HST”, 3. Select “@ssb” (Solar System Barycenter), 4. Select a date range that spans the days relevant to your observations. Then click Generate Ephemeris and click Download Results.
leapdir
Only used for HST analyses. The folder with respect to hst_cal where leapsecond calibration files will be saved.
flatfile
Only used for HST analyses. The path with respect to hst_cal to the flatfield file to use. The WFC3 flats can be downloaded here (G102) and here (G141); be sure to unzip the files after downloading them.
ywindow & xwindow
Can be set if one wants to remove edge effects (e.g.: many nans at the edges).
Below an example with the following setting:
ywindow [5,64]
xwindow [100,1700]
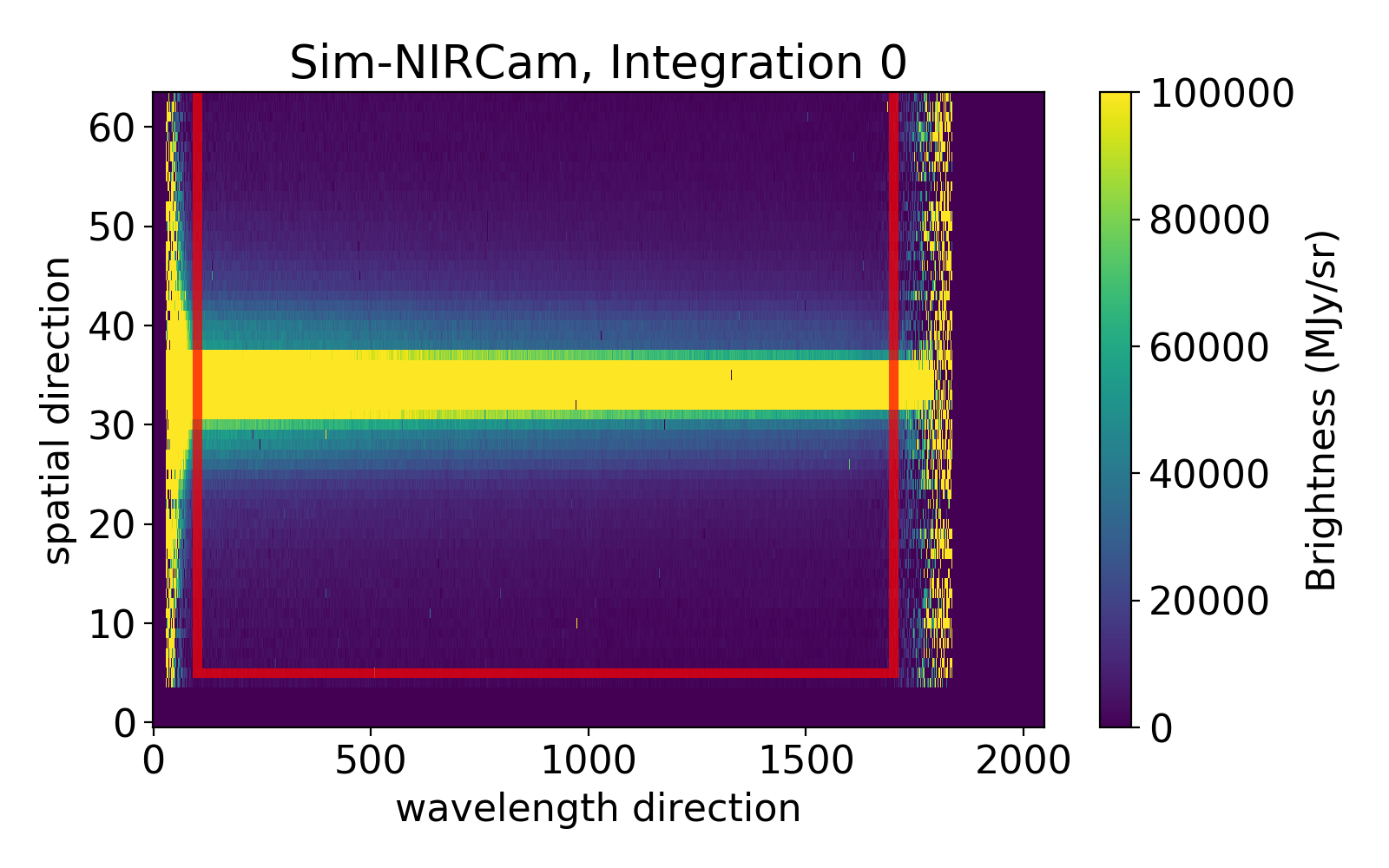
Everything outside of the box will be discarded and not used in the analysis.
src_pos_type
Determine the source position on the detector. Options: header, gaussian, weighted, max, or hst. The value ‘header’ uses the value of SRCYPOS in the FITS header.
record_ypos
Option to record the cross-dispersion trace position and width (if Gaussian fit) for each integration.
dqmask
Masks odd data quality (DQ) entries which indicate “Do not use” pixels following the jwst package documentation: https://jwst-pipeline.readthedocs.io/en/latest/jwst/references_general/references_general.html#data-quality-flags
expand
Super-sampling factor along cross-dispersion direction.
centroidtrim
Only used for HST analyses. The box width to cut around the centroid guess to perform centroiding on the direct images. This should be an integer.
centroidguess
Only used for HST analyses. A guess for the location of the star in the direct images in the format [x, y].
flatoffset
Only used for HST analyses. The positional offset to use for flatfielding. This should be formatted as a 2 element list with x and y offsets.
flatsigma
Only used for HST analyses. Used to sigma clip bad values from the flatfield image.
diffthresh
Only used for HST analyses. Sigma theshold for bad pixel identification in the differential non-destructive reads (NDRs).
bg_hw & spec_hw
bg_hw and spec_hw set the background and spectrum aperture relative to the source position.
Let’s looks at an example with the following settings:
bg_hw = 23
spec_hw = 18
Looking at the fits file science header, we can determine the source position:
src_xpos = hdulist['SCI',1].header['SRCXPOS']-xwindow[0]
src_ypos = hdulist['SCI',1].header['SRCYPOS']-ywindow[0]
Let’s assume in our example that src_ypos = 29.
(xwindow[0] and ywindow[0] corrects for the trimming of the data frame, as the edges were removed with the xwindow and ywindow parameters)
The plot below shows you which parts will be used for the background calculation (shaded in white; between the lower edge and src_ypos - bg_hw, and src_ypos + bg_hw and the upper edge) and which for the spectrum flux calculation (shaded in red; between src_ypos - spec_hw and src_ypos + spec_hw).
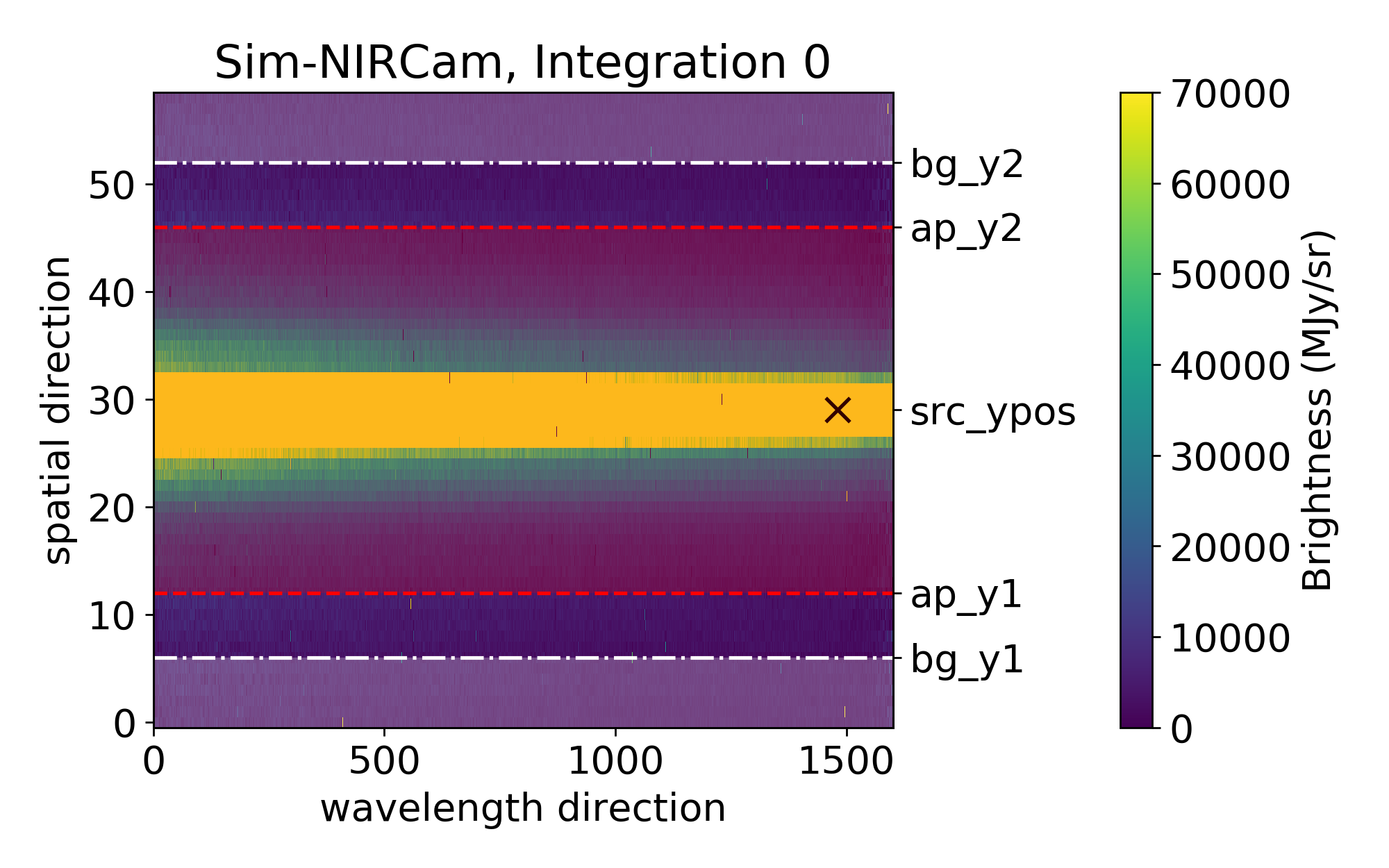
If you want to try multiple values sequentially, you can provide a list in the format [Start, Stop, Step]; this will give you sizes ranging from Start to Stop (inclusively) in steps of size Step. For example, [10,14,2] tries [10,12,14], but [10,15,2] still tries [10,12,14]. If spec_hw and bg_hw are both lists, all combinations of the two will be attempted.
ff_outlier
Set False to use only the background region when searching for outliers along the time axis (recommended for deep transits). Set True to apply the outlier rejection routine to the full frame (works well for shallow transits/eclipses). Be sure to check the percentage of pixels that were flagged while ff_outlier = True; the value should be << 1% when bg_thresh = [5,5].
bg_thresh
Double-iteration X-sigma threshold for outlier rejection along time axis.
The flux of every full-frame or background pixel will be considered over time for the current data segment.
e.g: bg_thresh = [5,5]: Two iterations of 5-sigma clipping will be performed in time for every full-frame or background pixel. Outliers will be masked and not considered in the flux calculation.
bg_deg
Sets the degree of the column-by-column background subtraction. If bg_deg is negative, use the median background of entire frame. Set to None for no background subtraction. Also, best to emphasize that we’re performing column-by-column BG subtraction
The function is defined in S3_data_reduction.optspex.fitbg
Possible values:
bg_deg = None: No backgound subtraction will be performed.bg_deg < 0: The median flux value in the background area will be calculated and subtracted from the entire 2D Frame for this paticular integration.bg_deg => 0: A polynomial of degree bg_deg will be fitted to every background column (background at a specific wavelength). If the background data has an outlier (or several) which is (are) greater than 5 * (Mean Absolute Deviation), this value will be not considered as part of the background. Step-by-step:
Take background pixels of first column
Fit a polynomial of degree
bg_degto the background pixels.Calculate the residuals (flux(bg_pixels) - polynomial_bg_deg(bg_pixels))
Calculate the MAD (Mean Absolute Deviation) of the greatest background outlier.
If MAD of the greatest background outlier is greater than 5, remove this background pixel from the background value calculation. Repeat from Step 2. and repeat as long as there is no 5*MAD outlier in the background column.
Calculate the flux of the polynomial of degree
bg_deg(calculated in Step 2) at the spectrum and subtract it.
bg_disp
Set True to perform row-by-row background subtraction (only useful for NIRCam).
bg_x1
Left edge of exclusion region for row-by-row background subtraction.
bg_x2
Right edge of exclusion region for row-by-row background subtraction.
p3thresh
Only important if bg_deg => 0 (see above). # sigma threshold for outlier rejection during background subtraction which corresponds to step 3 of optimal spectral extraction, as defined by Horne (1986).
p5thresh
Used during Optimal Extraction. # sigma threshold for outlier rejection during step 5 of optimal spectral extraction, as defined by Horne (1986). Default is 10. For more information, see the source code of optspex.optimize.
p7thresh
Used during Optimal Extraction. # sigma threshold for outlier rejection during step 7 of optimal spectral extraction, as defined by Horne (1986). Default is 10. For more information, see the source code of optspex.optimize.
fittype
Used during Optimal Extraction. fittype defines how to construct the normalized spatial profile for optimal spectral extraction. Options are: ‘smooth’, ‘meddata’, ‘wavelet’, ‘wavelet2D’, ‘gauss’, or ‘poly’. Using the median frame (meddata) should work well with JWST. Otherwise, using a smoothing function (smooth) is the most robust and versatile option. Default is meddata. For more information, see the source code of optspex.optimize.
window_len
Used during Optimal Extraction. window_len is only used when fittype = ‘smooth’ or ‘meddata’ (when computing median frame). It sets the length scale over which the data are smoothed. You can set this to 1 for no smoothing when computing median frame for fittype=meddata.
For more information, see the source code of optspex.optimize.
median_thresh
Used during Optimal Extraction. Sigma threshold when flagging outliers in median frame, when fittype=meddata and window_len > 1. Default is 5.
prof_deg
Used during Optimal Extraction. prof_deg is only used when fittype = ‘poly’. It sets the polynomial degree when constructing the spatial profile. Default is 3. For more information, see the source code of optspex.optimize.
iref
Only used for HST analyses. The file indices to use as reference frames for 2D drift correction. This should be a 1-2 element list with the reference indices for each scan direction.
curvature
Current options: ‘None’, ‘correct’. Using ‘None’ will not use any curvature correction and is strongly recommended against for instruments with strong curvature like NIRSpec/G395. Using ‘correct’ will bring the center of mass of each column to the center of the detector and perform the extraction on this straightened trace. If using ‘correct’, you should also be using fittype = ‘meddata’.
flag_bg
Only used for photometry analyses. Options are: True, False. Does an outlier rejection along the time axis for each individual pixel in a segment (= in a calints file).
interp_method
Only used for photometry analyses. Interpolate bad pixels. Options: None (if no interpolation should be performed), linear, nearest, cubic
centroid_method
Only used for photometry analyses. Selects the method used for determining the centroid position (options: fgc or mgmc). For it’s initial centroid guess, the ‘mgmc’ method creates a median frame from each batch of integrations and performs centroiding on the median frame (with the exact centroiding method set by the centroid_tech parameter). For each integration, the ‘mgmc’ method will then crop out an area around that guess using the value of ctr_cutout_size, and then perform a second round of centroiding to measure how the centroid moves over time. The ‘fgc’ method is the legacy centroiding method and is not currently recommended.
ctr_guess
Optional, and only used for photometry analyses. An initial guess for the [x, y] location of the star that will replace the default behavior of first doing a full-frame Gaussian centroiding to get an initial guess.
ctr_cutout_size
Only used for photometry analyses. For the ‘fgc’ and ‘mgmc’ methods this parameter is the amount of pixels all around the guessed centroid location which should be used for the more precise second centroid determination after the coarse centroid calculation. E.g., if ctr_cutout_size = 10 and the centroid (as determined after coarse step) is at (200, 200) then the cutout will have its corners at (190,190), (210,210), (190,210) and (210,190). The cutout therefore has the dimensions 21 x 21 with the centroid pixel (determined in the coarse centroiding step) in the middle of the cutout image.
oneoverf_corr
Only used for photometry analyses. The NIRCam detector exhibits 1/f noise along the long axis. Furthermore, each amplifier area (each is 512 colomns in length) has its own 1/f characteristics. Correcting for the 1/f effect will improve the quality of the final light curve. So, performing this correction is advised if it has not been done in any of the previous stages. The 1/f correction in Stage 3 treats every amplifier region separately. It does a row by row subtraction while avoiding pixels close to the star (see oneoverf_dist). “oneoverf_corr” sets which method should be used to determine the average flux value in each row of an amplifier region. Options: None, meanerr, median. If the user sets oneoverf_corr = None, no 1/f correction will be performed in S3. meanerr calculates a mean value which is weighted by the error array in a row. median calculated the median flux in a row.
oneoverf_dist
Only used for photometry analyses. Set how many pixels away from the centroid should be considered as background during the 1/f correction. E.g., Assume the frame has the shape 1000 in x and 200 in y. The centroid is at x,y = 400,100. Assume, oneoverf_dist has been set to 250. Then the area 0-150 and 650-1000 (in x) will be considered as background during the 1/f correction. The goal of oneoverf_dist is therefore basically to not subtract starlight during the 1/f correction.
skip_apphot_bg
Only used for photometry analyses. Skips the background subtraction in the aperture photometry routine. If the user does the 1/f noise subtraction during S3, the code will subtract the background from each amplifier region. The aperture photometry code will again subtract a background flux from the target flux by calculating the flux in an annulus in the background. If the user wants to skip this background subtraction by setting an background annulus, skip_apphot_bg has to be set to True.
photap
Only used for photometry analyses. Size of photometry aperture in pixels. The shape of the aperture is a circle. If the center of a pixel is not included within the aperture, it is being considered. If you want to try multiple values sequentially, you can provide a list in the format [Start, Stop, Step]; this will give you sizes ranging from Start to Stop (inclusively) in steps of size Step. For example, [10,14,2] tries [10,12,14], but [10,15,2] still tries [10,12,14]. If skyin and/or skywidth are also lists, all combinations of the three will be attempted.
skyin
Only used for photometry analyses. Inner sky annulus edge, in pixels. If you want to try multiple values sequentially, you can provide a list in the format [Start, Stop, Step]; this will give you sizes ranging from Start to Stop (inclusively) in steps of size Step. For example, [10,14,2] tries [10,12,14], but [10,15,2] still tries [10,12,14]. If photap and/or skywidth are also lists, all combinations of the three will be attempted.
skywidth
Only used for photometry analyses. The width of the sky annulus, in pixels. If you want to try multiple values sequentially, you can provide a list in the format [Start, Stop, Step]; this will give you sizes ranging from Start to Stop (inclusively) in steps of size Step. For example, [10,14,2] tries [10,12,14], but [10,15,2] still tries [10,12,14]. If photap and/or skyin are also lists, all combinations of the three will be attempted.
centroid_tech
Only used for photometry analyses. The centroiding technique used if centroid_method is set to mgmc. The options are: com, 1dg, 2dg. The recommended technique is com (standing for Center of Mass). More details about the options can be found in the photutils documentation at https://photutils.readthedocs.io/en/stable/centroids.html.
gauss_frame
Only used for photometry analyses. Range away from first centroid guess to include in centroiding map for gaussian widths. Only required for mgmc method. Options: 1 -> Max frame size (type integer).
isplots_S3
Sets how many plots should be saved when running Stage 3. A full description of these outputs is available here: Stage 3 Output
nplots
Sets how many integrations will be used for per-integration figures (Figs 3301, 3302, 3303, 3307, 3501, 3505). Useful for in-depth diagnoses of a few integrations without making thousands of figures. If set to None, a plot will be made for every integration.
vmin
Optional. Sets the vmin of the color bar for Figure 3101. Defaults to 0.97.
vmax
Optional. Sets the vmax of the color bar for Figure 3101. Defaults to 1.03.
time_axis
Optional. Determines whether the time axis in Figure 3101 is along the y-axis (‘y’) or the x-axis (‘x’). Defaults to ‘y’.
testing_S3
If set to True only the last segment (which is usually the smallest) in the inputdir will be run. Also, only five integrations from the last segment will be reduced.
save_output
If set to True output will be saved as files for use in S4. Setting this to False is useful for quick testing
save_fluxdata
If set to True (the default if save_fluxdata is not in your ECF), then save FluxData outputs for debugging or use with other tools. Note that these can be quite large files and may fill your drive if you are trying many spec_hw,bg_hw pairs.
hide_plots
If True, plots will automatically be closed rather than popping up on the screen.
verbose
If True, more details will be printed about steps.
topdir + inputdir
The path to the directory containing the Stage 2 JWST data, or, for HST observations, the _ima FITS files (including both direct images and spectra) downloaded from MAST. Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 3 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
topdir + time_file
Optional. The path to a file that contains the time array you want to use instead of the one contained in the FITS file. Directories containing spaces should be enclosed in quotation marks.
Stage 4
# Eureka! Control File for Stage 4: Generate Lightcurves
# Stage 4 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-4
# Number of spectroscopic channels spread evenly over given wavelength range
nspecchan 2 # Number of spectroscopic channels spread evenly over given wavelength range. Set to None to leave the spectrum unbinned.
compute_white True # Also compute the white-light lightcurve
wave_min 1.5 # Minimum wavelength. Set to None to use the shortest extracted wavelength from Stage 3.
wave_max 4.5 # Maximum wavelength. Set to None to use the longest extracted wavelength from Stage 3.
allapers False # Run S4 on all of the apertures considered in S3? Otherwise will use newest output in the inputdir
# Manualy mask pixel columns by index number
# mask_columns []
# Parameters for drift correction of 1D spectra
recordDrift False # Set True to record drift/jitter in 1D spectra (always recorded if correctDrift is True)
correctDrift False # Set True to correct drift/jitter in 1D spectra (not recommended for simulated data)
drift_preclip 0 # Ignore first drift_preclip points of spectrum
drift_postclip 100 # Ignore last drift_postclip points of spectrum, None = no clipping
drift_range 11 # Trim spectra by +/-X pixels to compute valid region of cross correlation
drift_hw 5 # Half-width in pixels used when fitting Gaussian, must be smaller than drift_range
drift_iref -1 # Index of reference spectrum used for cross correlation, -1 = last spectrum
sub_mean True # Set True to subtract spectrum mean during cross correlation
sub_continuum True # Set True to subtract the continuum from the spectra using a highpass filter
highpassWidth 10 # The integer width of the highpass filter when subtracting the continuum
# Parameters for sigma clipping
clip_unbinned False # Whether or not sigma clipping should be performed on the unbinned 1D time series
clip_binned True # Whether or not sigma clipping should be performed on the binned 1D time series
sigma 10 # The number of sigmas a point must be from the rolling median to be considered an outlier
box_width 10 # The width of the box-car filter (used to calculated the rolling median) in units of number of data points
maxiters 5 # The number of iterations of sigma clipping that should be performed.
boundary 'fill' # Use 'fill' to extend the boundary values by the median of all data points (recommended), 'wrap' to use a periodic boundary, or 'extend' to use the first/last data points
fill_value mask # Either the string 'mask' to mask the outlier values (recommended), 'boxcar' to replace data with the mean from the box-car filter, or a constant float-type fill value.
# Limb-darkening parameters needed to compute exotic-ld
compute_ld True
inst_filter prism # Filter of JWST/HST instrument, supported list see https://exotic-ld.readthedocs.io/en/latest/views/supported_instruments.html
metallicity 0.1 # Metallicity of the star
teff 6000 # Effective temperature of the star in K
logg 4.0 # Surface gravity in log g
exotic_ld_direc /home/User/exotic-ld_data/ # Directory for ancillary files for exotic-ld, download from: https://zenodo.org/record/6344946
exotic_ld_grid 3D # 1D or 3D model grid
exotic_ld_file /home/User/exotic-ld_data/Custom_throughput_file.csv # Custom throughput file, for examples see Eureka/demos/JWST/Custom_throughput_files
# Diagnostics
isplots_S4 3 # Generate few (1), some (3), or many (5) figures (Options: 1 - 5)
vmin 0.97 # Sets the vmin of the color bar for Figure 4101.
vmax 1.03 # Sets the vmax of the color bar for Figure 4101.
time_axis 'y' # Determines whether the time axis in Figure 4101 is along the y-axis ('y') or the x-axis ('x')
hide_plots False # If True, plots will automatically be closed rather than popping up
verbose True # If True, more details will be printed about steps
# Project directory
topdir /home/User/Data/JWST-Sim/NIRSpec/
# Directories relative to topdir
inputdir Stage3 # The folder containing the outputs from Eureka!'s S3 or JWST's S3 pipeline (will be overwritten if calling S3 and S4 sequentially)
outputdir Stage4
nspecchan
Number of spectroscopic channels spread evenly over given wavelength range. Set to None to leave the spectrum unbinned.
compute_white
If True, also compute the white-light lightcurve.
wave_min & wave_max
Start and End of the wavelength range being considered. Set to None to use the shortest/longest extracted wavelength from Stage 3.
allapers
If True, run S4 on all of the apertures considered in S3. Otherwise the code will use the only or newest S3 outputs found in the inputdir. To specify a particular S3 save file, ensure that “inputdir” points to the procedurally generated folder containing that save file (e.g. set inputdir to /Data/JWST-Sim/NIRCam/Stage3/S3_2021-11-08_nircam_wfss_ap10_bg10_run1/).
mask_columns
List of pixel columns that should not be used when constructing a light curve. Absolute (not relative) pixel columns should be used. Figure 3102 is very helpful for identifying bad pixel columns.
recordDrift
If True, compute drift/jitter in 1D spectra (always recorded if correctDrift is True)
correctDrift
If True, correct for drift/jitter in 1D spectra.
drift_preclip
Ignore first drift_preclip points of spectrum when correcting for drift/jitter in 1D spectra.
drift_postclip
Ignore the last drift_postclip points of spectrum when correcting for drift/jitter in 1D spectra. None = no clipping.
drift_range
Trim spectra by +/- drift_range pixels to compute valid region of cross correlation when correcting for drift/jitter in 1D spectra.
drift_hw
Half-width in pixels used when fitting Gaussian when correcting for drift/jitter in 1D spectra. Must be smaller than drift_range.
drift_iref
Index of reference spectrum used for cross correlation when correcting for drift/jitter in 1D spectra. -1 = last spectrum.
sub_mean
If True, subtract spectrum mean during cross correlation (can help with cross-correlation step).
sub_continuum
Set True to subtract the continuum from the spectra using a highpass filter
highpassWidth
The integer width of the highpass filter when subtracting the continuum
clip_unbinned
Whether or not sigma clipping should be performed on the unbinned 1D time series
clip_binned
Whether or not sigma clipping should be performed on the binned 1D time series
sigma
Only used if sigma_clip=True. The number of sigmas a point must be from the rolling median to be considered an outlier
box_width
Only used if sigma_clip=True. The width of the box-car filter (used to calculated the rolling median) in units of number of data points
maxiters
Only used if sigma_clip=True. The number of iterations of sigma clipping that should be performed.
boundary
Only used if sigma_clip=True. Use ‘fill’ to extend the boundary values by the median of all data points (recommended), ‘wrap’ to use a periodic boundary, or ‘extend’ to use the first/last data points
fill_value
Only used if sigma_clip=True. Either the string ‘mask’ to mask the outlier values (recommended), ‘boxcar’ to replace data with the mean from the box-car filter, or a constant float-type fill value.
sum_reads
Only used for HST analyses. Should differential non-destructive reads be summed together to reduce noise and data volume or not.
compute_ld
Whether or not to compute limb-darkening coefficients using exotic-ld.
inst_filter
Used by exotic-ld if compute_ld=True. The filter of JWST/HST instrument, supported list see https://exotic-ld.readthedocs.io/en/latest/views/supported_instruments.html (leave off the observatory and instrument so that JWST_NIRSpec_Prism becomes just Prism).
metallicity
Used by exotic-ld if compute_ld=True. The metallicity of the star.
teff
Used by exotic-ld if compute_ld=True. The effective temperature of the star in K.
logg
Used by exotic-ld if compute_ld=True. The surface gravity in log g.
exotic_ld_direc
Used by exotic-ld if compute_ld=True. The fully qualified path to the directory for ancillary files for exotic-ld, download at https://zenodo.org/record/6344946.
exotic_ld_grid
Used by exotic-ld if compute_ld=True. 1D or 3D model grid.
exotic_ld_file
Used by exotic-ld as throughput input file. If none, exotic-ld uses throughput from ancillary files. Make sure that wavelength is given in Angstrom!
isplots_S4
Sets how many plots should be saved when running Stage 4. A full description of these outputs is available here: Stage 4 Output
nplots
Sets how many integrations will be used for per-integration figures (Figs 4301 and 4302). Useful for in-depth diagnoses of a few integrations without making thousands of figures. If set to None, a plot will be made for every integration.
vmin
Optional. Sets the vmin of the color bar for Figure 4101. Defaults to 0.97.
vmax
Optional. Sets the vmax of the color bar for Figure 4101. Defaults to 1.03.
time_axis
Optional. Determines whether the time axis in Figure 4101 is along the y-axis (‘y’) or the x-axis (‘x’). Defaults to ‘y’.
hide_plots
If True, plots will automatically be closed rather than popping up on the screen.
verbose
If True, more details will be printed about steps.
topdir + inputdir
The path to the directory containing the Stage 3 JWST data. Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 4 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
Stage 5
# Eureka! Control File for Stage 5: Lightcurve Fitting
# Stage 5 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-5
ncpu 1 # The number of CPU threads to use when running emcee or dynesty in parallel
allapers False # Run S5 on all of the apertures considered in S4? Otherwise will use newest output in the inputdir
rescale_err False # Rescale uncertainties to have reduced chi-squared of unity
fit_par ./S5_fit_par_template.epf # What fitting epf do you want to use?
verbose True # If True, more details will be printed about steps
fit_method [dynesty] #options are: lsq, emcee, dynesty (can list multiple types separated by commas)
run_myfuncs [batman_tr, polynomial] # options are: batman_tr, batman_ecl, sinusoid_pc, expramp, hstramp, polynomial, step, xpos, ypos, xwidth, ywidth, and GP (can list multiple types separated by commas)
# Manual clipping in time
manual_clip None # A list of lists specifying the start and end integration numbers for manual removal (done before summing reads).
# Limb darkening controls
# IMPORTANT: limb-darkening coefficients are not automatically fixed then, change to 'fixed' in .epf file whether they should be fixed or fitted!
use_generate_ld None # use the generated limb-darkening coefficients from Stage 4? Options: exotic-ld, None. For exotic-ld, the limb-darkening laws available are linear, quadratic, 3-parameter and 4-parameter non-linear.
ld_file None # Fully qualified path to the location of a limb darkening file that you want to use
ld_file_white None # Fully qualified path to the location of a limb darkening file that you want to use for the white-light light curve (required if ld_file is not None and any EPF parameters are set to white_free or white_fixed).
# General fitter
old_fitparams None # filename relative to topdir that points to a fitparams csv to resume where you left off (set to None to start from scratch)
#lsq
lsq_method 'Nelder-Mead' # The scipy.optimize.minimize optimization method to use
lsq_tol 1e-6 # The tolerance for the scipy.optimize.minimize optimization method
lsq_maxiter None # Maximum number of iterations to perform. Depending on the method each iteration may use several function evaluations. Set to None to use the default value
#mcmc
old_chain None # Output folder relative to topdir that contains an old emcee chain to resume where you left off (set to None to start from scratch)
lsq_first True # Initialize with an initial lsq call (can help shorten burn-in, but turn off if lsq fails). Only used if old_chain is None
run_nsteps 1000
run_nwalkers 200
run_nburn 500 # How many of run_nsteps should be discarded as burn-in steps
#dynesty
run_nlive 1024 # Must be > ndim * (ndim + 1) // 2
run_bound 'multi'
run_sample 'auto'
run_tol 0.1
#GP inputs
kernel_inputs ['time'] #options: time
kernel_class ['Matern32'] #options: ExpSquared, Matern32, Exp, RationalQuadratic for george, Matern32 for celerite (sums of kernels possible for george separated by commas)
GP_package 'celerite' #options: george, celerite
useHODLR False # Set to True to use the HODLRSolver when using the george GP
# Plotting controls
interp False # Should astrophysical model be interpolated (useful for uneven sampling like that from HST)
# Diagnostics
isplots_S5 5 # Generate few (1), some (3), or many (5) figures (Options: 1 - 5)
nbin_plot 100 # The number of bins that should be used for figures 5104 and 5304. Defaults to 100.
testing_S5 False # Boolean, set True to only use the first spectral channel
testing_model False # Boolean, set True to only inject a model source of systematics
hide_plots False # If True, plots will automatically be closed rather than popping up
# Project directory
topdir /home/User/Data/JWST-Sim/NIRSpec/
# Directories relative to topdir
inputdir Stage4 # The folder containing the outputs from Eureka!'s S4 pipeline (will be overwritten if calling S4 and S5 sequentially)
outputdir Stage5
ncpu
Integer. Sets the number of CPUs to use for multiprocessing Stage 5 fitting.
allapers
Boolean to determine whether Stage 5 is run on all the apertures considered in Stage 4. If False, will just use the most recent output in the input directory.
rescale_err
Boolean to determine whether the uncertainties will be rescaled to have a reduced chi-squared of 1
fit_par
Path to Stage 5 priors and fit parameter file.
verbose
If True, more details will be printed about steps.
fit_method
Fitting routines to run for Stage 5 lightcurve fitting. For standard numpy functions, this can be one or more of the following: [lsq, emcee, dynesty]. For theano-based differentiable functions, this can be one or more of the following: [exoplanet, nuts] where exoplanet uses a gradient based optimization method and nuts uses the No U-Turn Sampling method implemented in PyMC3.
run_myfuncs
Determines the astrophysical and systematics models used in the Stage 5 fitting. For standard numpy functions, this can be one or more (separated by commas) of the following: [batman_tr, batman_ecl, sinusoid_pc, expramp, hstramp, polynomial, step, xpos, ypos, xwidth, ywidth, GP]. For theano-based differentiable functions, this can be one or more of the following: [starry, sinusoid_pc, expramp, hstramp, polynomial, step, xpos, ypos, xwidth, ywidth], where starry replaces both the batman_tr and batman_ecl models and offers a more complicated phase variation model than sinusoid_pc that accounts for eclipse mapping signals.
manual_clip
Optional. A list of lists specifying the start and end integration numbers for manual removal. E.g., to remove the first 20 data points specify [[0,20]], and to also remove the last 20 data points specify [[0,20],[-20,None]]. If you want to clip the 10th integration, this would be index 9 since python uses zero-indexing. And the manual_clip start and end values are used to slice a numpy array, so they follow the same convention of inclusive start index and exclusive end index. In other words, to trim the 10th integrations, you would set manual_clip to [[9,10]].
Limb Darkening Parameters
The following three parameters control the use of pre-generated limb darkening coefficients.
use_generate_ld
If you want to use the generated limb-darkening coefficients from Stage 4, use exotic-ld. Otherwise, use None. Important: limb-darkening coefficients are not automatically fixed, change the limb darkening parameters to ‘fixed’ in the .epf file if they should be fixed instead of fitted! The limb-darkening laws available to exotic-ld are linear, quadratic, 3-parameter and 4-parameter non-linear.
ld_file
If you want to use custom calculated limb-darkening coefficients, set to the fully qualified path to a file containing limb darkening coefficients that you want to use. Otherwise, set to None. Note: this option only works if use_generate_ld=None. The file should be a plain .txt file with one column for each limb darkening coefficient and one row for each wavelength range.
ld_file_white
The same type of parameter as ld_file, but for the limb-darkening coefficients to be used for the white-light fit. This parameter is required if ld_file is not None and any of your EPF parameters are set to white_free or white_fixed. If no parameter is set to white_free or white_fixed, then this parameter is ignored.
GP parameters
The following parameters control part of the GP model (if listed in run_myfuncs).
kernel_inputs
Only used for fits with a GP. A list of the covariates to be used when fitting a GP to the model. At present, only GPs as a function of time are allowed, so this must be [‘time’]
kernel_class
Only used for fits with a GP. A list of the types of GP kernels to use. For the george GP package, this includes ExpSquared, Matern32, Exp, and RationalQuadratic. For the celerite package, this only includes Matern32. It is possible to sum multiple kernels possible for george by listing multiple kernels.
GP_package
Only used for fits with a GP. The Python GP package to use, with the options of ‘george’ or ‘celerite’.
useHODLR
Only used for fits with a GP. If True and GP_package is set to ‘george’, use the (potentially faster) HODLRSolver instead of the (more robust) BasicSolver.
Least-Squares Fitting Parameters
The following set the parameters for running the least-squares fitter.
lsq_method
Least-squares fitting method: one of any of the scipy.optimize.minimize least-squares methods.
lsq_tolerance
Float to determine the tolerance of the scipy.optimize.minimize method.
Emcee Fitting Parameters
The following set the parameters for running emcee.
old_chain
Output folder containing previous emcee chains to resume previous runs. To start from scratch, set to None.
lsq_first
Boolean to determine whether to run least-squares fitting before MCMC. This can shorten burn-in but should be turned off if least-squares fails. Only used if old_chain is None.
run_nsteps
Integer. The number of steps for emcee to run.
run_nwalkers
Integer. The number of walkers to use.
run_nburn
Integer. The number of burn-in steps to run.
Dynesty Fitting Parameters
The following set the parameters for running dynesty. These options are described in more detail in: https://dynesty.readthedocs.io/en/latest/api.html?highlight=unif#module-dynesty.dynesty
run_nlive
Integer. Number of live points for dynesty to use. Should be at least greater than (ndim * (ndim+1)) / 2, where ndim is the total number of fitted parameters. For shared fits, multiply the number of free parameters by the number of wavelength bins specified in Stage 4. For convenience, this can be set to ‘min’ to automatically set run_nlive to (ndim * (ndim+1)) / 2.
run_bound
The bounding method to use. Options are: [‘none’, ‘single’, ‘multi’, ‘balls’, ‘cubes’]
run_sample
The sampling method to use. Options are [‘auto’, ‘unif’, ‘rwalk’, ‘rstagger’, ‘slice’, ‘rslice’, ‘hslice’]
run_tol
Float. The tolerance for the dynesty run. Determines the stopping criterion. The run will stop when the estimated contribution of the remaining prior volume to the total evidence falls below this threshold.
NUTS Fitting Parameters
The following set the parameters for running PyMC3’s NUTS sampler. These options are described in more detail in: https://docs.pymc.io/en/v3/api/inference.html#pymc3.sampling.sample
tune
Number of iterations to tune. Samplers adjust the step sizes, scalings or similar during tuning. Tuning samples will be drawn in addition to the number specified in the draws argument.
draws
The number of samples to draw. The number of tuned samples are discarded by default.
chains
The number of chains to sample. Running independent chains is important for some convergence statistics and can also reveal multiple modes in the posterior. If None, then set to either ncpu or 2, whichever is larger.
target_accept
Adapt the step size such that the average acceptance probability across the trajectories are close to target_accept. Higher values for target_accept lead to smaller step sizes. A default of 0.8 is recommended, but setting this to higher values like 0.9 or 0.99 can help with sampling from difficult posteriors. Valid values are between 0 and 1 (exclusive).
force_positivity
Used by the sinusoid_pc model. If True, force positive phase variations (phase variations that never go below the bottom of the eclipse). Physically speaking, a negative phase curve is impossible, but strictly enforcing this can hide issues with the decorrelation or potentially bias your measured minimum flux level. Either way, use caution when choosing the value of this parameter.
interp
Boolean to determine whether the astrophysical model is interpolated when plotted. This is useful when there is uneven sampling in the observed data.
isplots_S5
Sets how many plots should be saved when running Stage 5. A full description of these outputs is available here: Stage 5 Output
nbin_plot
The number of bins that should be used for figures 5104 and 5304. Defaults to 100.
hide_plots
If True, plots will automatically be closed rather than popping up on the screen.
topdir + inputdir
The path to the directory containing the Stage 4 JWST data. Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 5 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
Stage 5 Fit Parameters
Warning
The Stage 5 fit parameter file has the file extension .epf, not .ecf. These have different formats, and are not interchangeable.
This file describes the transit/eclipse and systematics parameters and their prior distributions. Each line describes a new parameter, with the following basic format:
Name Value Free PriorPar1 PriorPar2 PriorType
Namedefines the specific parameter being fit for. Available options are:- Transit and Eclipse Parameters
rp- planet-to-star radius ratio, for the transit models.fp- planet-to-star flux ratio, for the eclipse models.
- Orbital Parameters
per- orbital period (in days)t0- transit time (in the same units as your input data - most likely BMJD_TDB)time_offset- (optional), the absolute time offset of your time-series data (in days)inc- orbital inclination (in degrees)a- a/R*, the ratio of the semimajor axis to the stellar radiusecc- orbital eccentricityw- argument of periapsis (degrees)Rs- the host star’s radius in units of solar radii.This parameter is recommended for batman_ecl fits as it allows for a conversion of a/R* to physical units in order to account for light travel time. If not provided for batman_ecl fits, the finite speed of light will not be accounted for. Fits with the starry model require that
Rsbe provided as starry always accounts for light travel time. This parameter should be set tofixedunless you really want to marginalize overRs.Ms- the host star’s mass in units of solar masses.This parameter is required for fits with the starry model as starry currently requires the parameter to be provided. In practice, the stellar mass is not actually used in Eureka! though as we allow for
aandperto be provided directly. This parameter should be set tofixedunless you really want to marginalize overMs.
- Sinusoidal Phase Curve Parameters
The sinusoid_pc phase curve model for the standard numpy models allows for the inclusion of up to four sinusoids into a single phase curve. The theano-based differentiable functions allow for any number of sinusoids.
AmpCos1- Amplitude of the first cosine with one peak near eclipse (orbital phase 0.5)AmpSin1- Amplitude of the first sine with one peak near quadrature at orbital phase 0.75AmpCos2- Amplitude of the second cosine with two peaks near eclipse (orbital phase 0.5) and transit (orbital phase 0)AmpSin2- Amplitude of the second sine with two peaks near quadrature at orbital phases 0.25 and 0.75
- Starry Phase Curve and Eclipse Mapping Parameters
The starry model allows for the modelling of an arbitrarily complex phase curve by fitting the phase curve using spherical harmonics terms for the planet’s brightness map
Yl_m- Spherical harmonic coefficients normalized by the Y0_0 term wherelandmshould be replaced with integers.lcan be any integer greater than or equal to 1, andmcan be any integer between-lto+l. For example, theY1_0term fits for the sub-stellar to anti-stellar brightness ratio (comparable toAmpCos1), theY1_1term fits for the East–West brightness ratio (comparable to-AmpSin1), and theY1_-1term fits for the North–South pole brightness ratio (undetectable using phase variations, but potentially detectable using eclipse mapping). TheY0_0term cannot be fit directly but is instead fit through the more observablefpterm which is composed of theY0_0term and the square of therpterm.
- Limb Darkening Parameters
limb_dark- The limb darkening model to be used.Options are:
['uniform', 'linear', 'quadratic', 'kipping2013', 'squareroot', 'logarithmic', 'exponential', '4-parameter'].uniformlimb-darkening has no parameters,linearhas a single parameteru1,quadratic,kipping2013,squareroot,logarithmic, andexponentialhave two parametersu1, u2, and4-parameterhas four parametersu1, u2, u3, u4.
- Systematics Parameters. Depends on the model specified in the Stage 5 ECF.
c0--c9- Coefficients for 0th to 3rd order polynomials.The polynomial coefficients are numbered as increasing powers (i.e.
c0a constant,c1linear, etc.). The x-values of the polynomial are the time with respect to the mean of the time of the lightcurve time array. Polynomial fits should include at leastc0for usable results.r0--r2andr3--r5- Coefficients for the first and second exponential ramp models.The exponential ramp model is defined as follows:
r0*np.exp(-r1*time_local + r2) + r3*np.exp(-r4*time_local + r5) + 1, wherer0--r2describe the first ramp, andr3--r5the second.time_localis the time relative to the first frame of the dataset. If you only want to fit a single ramp, you can omitr3--r5or set them as fixed to0. Users should not fit all three parameters from each model at the same time as there are significant degeneracies between ther0andr2parameters (andr3andr5parameters). One option is to setr0to the sign of the ramp (-1 for decaying, 1 for rising) while fitting for the remaining coefficients. Another option is to fit forr0--r1and setr2to zero. This option works well when fitting spectroscopic light curves that could be rising or decaying.h0--h5- Coefficients for the HST exponential + polynomial ramp model.The HST ramp model is defined as follows:
1 + h0*np.exp(-h1*time_batch + h2) + h3*time_batch + h4*time_batch**2, whereh0--h2describe the exponential ramp per HST orbit,h3--h4describe the polynomial (up to order two) per HST orbit, andh5is the orbital period of HST (in the same time units as the data, usually days). A good starting point forh5is 0.066422 days.time_batch = time_local % h5. If you want to fit a linear trend in time, you can omith4or fix it to0. Users should not fit all three parameters from the exponential model at the same time as there are significant degeneracies between theh0andh2parameters. One option is to seth0to the sign of the ramp (-1 for decaying, 1 for rising) while fitting for the remaining coefficients. Another option is to fit forh0--h1and seth2to zero. This option works well when fitting spectroscopic light curves that could be rising or decaying.step0andsteptime0- The step size and time for the first step function (useful for removing mirror segment tilt events).For additional steps, simply increment the integer at the end (e.g.
step1andsteptime1). The change in flux is relative to the flux from the previous step.xpos- Coefficient for linear decorrelation against drift/jitter in the x direction (spectral direction for spectroscopy data).xwidth- Coefficient for linear decorrelation against changes in the PSF width in the x direction (cross-correlation width in the spectral direction for spectroscopy data).ypos- Coefficient for linear decorrelation against drift/jitter in the y direction (spatial direction for spectroscopy data).ywidth- Coefficient for linear decorrelation against changes in the PSF width in the y direction (spatial direction for spectroscopy data).Aandm- The natural logarithm (ln) of the covariance amplitude and lengthscale to use for the GP model specified in your Stage 5 ECF.Significant care should be used when specifying the priors on these parameters as an excessively flexible GP model may end up competing with your astrophysical model. That said, there are no hard and fast rules about what your priors should be, and you will need to experiment to find what works best. If there are multiple kernels that are being added, the second kernel’s parameters will be
A1andm1, and so on.
White Noise Parameters - options are
scatter_multfor a multiplier to the expected noise from Stage 3 (recommended), orscatter_ppmto directly fit the noise level in ppm.
Free determines whether the parameter is fixed, free, white_fixed, white_free, independent, or shared.
fixed parameters are fixed in the fitting routine and not fit for.
free parameters are fit for according to the specified prior distribution, independently for each wavelength channel.
white_fixed and white_free parameters are first fit using free on the white-light light curve to update the prior for the parameter, and then treated as fixed or free (respectively) for the spectral fits.
shared parameters are fit for according to the specified prior distribution, but are common to all wavelength channels.
independent variables set auxiliary functions needed for the fitting routines.
The PriorType can be U (Uniform), LU (Log Uniform), or N (Normal). If U/LU, then PriorPar1 and PriorPar2 are the lower and upper limits of the prior distribution. If N, then PriorPar1 is the mean and PriorPar2 is the stadard deviation of the Gaussian prior.
Here’s an example fit parameter file:
# Stage 5 Fit Parameters Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-5-fit-parameters
# Name Value Free? PriorPar1 PriorPar2 PriorType
# "Free?" can be free, fixed, white_free, white_fixed, shared, or independent
# PriorType can be U (Uniform), LU (Log Uniform), or N (Normal).
# If U/LU, PriorPar1 and PriorPar2 represent upper and lower limits of the parameter/log(the parameter).
# If N, PriorPar1 is the mean and PriorPar2 is the standard deviation of a Gaussian prior.
#-------------------------------------------------------------------------------------------------------
#
# ------------------
# ** Transit/eclipse parameters **
# ------------------
rp 0.148 'free' 0.15 0.1 N
#fp 0.008 'free' 0 0.5 U
# ----------------------
# ** Phase curve parameters **
# ----------------------
#AmpCos1 0.4 'free' 0 1 U
#AmpSin1 0.01 'free' -1 1 U
#AmpCos2 0.01 'free' -1 1 U
#AmpSin2 0.01 'free' -1 1 U
# ------------------
# ** Orbital parameters **
# ------------------
per 4.055259 'free' 4.055259 1e-5 N
t0 57394.524 'free' 57394.524 0.05 N
time_offset 0 'independent'
inc 87.83 'free' 87.83 0.25 N
a 10.9 'free' 10.9 1 N # An incorrect a/R* seems to have been used in the simulations
ecc 0.0 'fixed' 0 1 U
w 90. 'fixed' 0 180 U
# -------------------------
# ** Limb darkening parameters **
# Choose limb_dark from ['uniform', 'linear', 'quadratic', 'kipping2013', 'squareroot', 'logarithmic', 'exponential','3-parameter', '4-parameter']
# When using generated limb-darkening coefficients from exotic-ld choose from ['linear', 'quadratic', '3-parameter', '4-parameter']
# -------------------------
limb_dark 'kipping2013' 'independent'
u1 0.59 'free' 0 1 U
u2 0.84 'free' 0 1 U
# --------------------
# ** Systematic variables **
# Polynomial model variables (c0--c9 for 0th--3rd order polynomials in time); Fitting at least c0 is very strongly recommended!
# Exponential ramp model variables (r0--r2 for one exponential ramp, r3--r5 for a second exponential ramp)
# HST exponential ramp + polynomial model variables (h0--h2 for exponential, h3--h4 for polynomial in time)
# GP model parameters (A, m for the first kernel; A1, m1 for the second kernel; etc.) in log scale
# Step-function model variables (step# and steptime# for step-function model #; e.g. step0 and steptime0)
# Drift model variables (xpos, ypos, xwidth, ywidth)
# --------------------
c0 1.015 'free' 1 0.05 N
c1 0 'free' 0 0.01 N
# A -7 'free' -15 0 U
# m -4 'free' -6 0 U
# -----------
# ** White noise **
# Use scatter_mult to fit a multiplier to the expected noise level from Stage 3 (recommended)
# Use scatter_ppm to fit the noise level in ppm
# -----------
scatter_mult 1.1 'free' 1.1 1 N
Stage 6
# Eureka! Control File for Stage 6: Spectra Plotting
# Stage 6 Documentation: https://eurekadocs.readthedocs.io/en/latest/ecf.html#stage-6
allapers False # Run S6 on all of the apertures considered in S5? Otherwise will use newest output in the inputdir
# Plotting parameters
y_params ['rp^2', 'u1', 'u2', 'c0', 'c1', 'scatter_mult'] # The parameter name as entered in your EPF file in Stage 5. rp^2, 1/r1, and 1/r4 are also permitted.
y_labels [None, None, None, None, None, None] # The formatted string you want on the y-label. Set to None to use the default formatting.
y_label_units [None, None, None, None, None, None] # The formatted string for the units you want on the y-label - e.g., (ppm), (seconds), '(days$^{-1})$', etc.. Set to None to automatically add any implied units from y_scalars (e.g. ppm), or set to '' to force no units.
y_scalars [1e6, 1, 1, 1, 1, 1] # Can be used to convert to percent (100), ppm (1e6), etc.
x_unit um # Options include any measurement of light included in astropy.units.spectral (e.g. um, nm, Hz, etc.)
# Tabulating parameters
ncol 4 # The number of columns you want in your LaTeX formatted tables
# This section is relevant if isplots_S6>=3
# Scale height parameters (if you want a second copy of the plot with a second y-axis with units of scale height)
star_Rad 0.6506 # The radius of the star in units of solar radii
planet_Teq 1400 # The equilibrium temperature of the planet in units of Kelvin
planet_Mass 2 # The planet's mass in units of Jupiter masses (used to calculate surface gravity)
planet_Rad None # The planet's radius in units of Jupiter radii (used to calculate surface gravity); Set to None to use the average fitted radius
planet_mu 2.3 # The mean molecular mass of the atmosphere (in atomic mass units)
planet_R0 None # The reference radius (in Jupiter radii) for the scale height measurement; Set to None to use the mean fitted radius
# Diagnostics
isplots_S6 5 # Generate few (1), some (3), or many (5) figures (Options: 1 - 5)
hide_plots False # If True, plots will automatically be closed rather than popping up
# Project directory
topdir /home/User/Data/JWST-Sim/NIRSpec/
# Model to plot underneath the fitted data points
model_spectrum None # Path to the model spectrum relative to topdir. Set to None if no model should be plotted.
model_x_unit um # Options include any measurement of light included in astropy.units.spectral (e.g. um, nm, Hz, etc.)
model_y_param rp^2 # Follow the same format as y_params. If desired, can be rp if y_params is rp^2, or vice-versa.
model_y_scalar 1 # Indicate whether model y-values have already been scaled (e.g. write 1e6 if model_spectrum is in ppm)
model_zorder 0 # The zorder of the model on the plot (0 for beneath the data, 1 for above the data)
model_delimiter None # Delimiter between columns. Typical options: None (for space), ',' for comma
# Directories relative to topdir
inputdir Stage5 # The folder containing the outputs from Eureka!'s S5 pipeline (will be overwritten if calling S5 and S6 sequentially)
outputdir Stage6
allapers
Boolean to determine whether Stage 6 is run on all the apertures considered in Stage 5. If False, will just use the most recent output in the input directory.
y_params
The parameter to use when plotting and saving the output table. To plot the transmission spectrum, the value can be ‘rp’ or ‘rp^2’. To plot the dayside emission spectrum, the value must be fp. To plot the spectral dependence of any other parameters, simply enter their name as formatted in your EPF. For convenience, it is also possible to plot ‘1/r1’ and ‘1/r4’ to visualize the exonential ramp timescales. It is also possible to plot ‘fn’ (the nightside flux from a sinusoidal phase curve), ‘pc_offset’ (the sinusoidal offset of the phase curve), ‘pc_amp’ (the sinusoidal amplitude of the phase curve), ‘pc_offset2’ (the second order sinusoidal offset of the phase curve), and ‘pc_amp2’ (the second order sinusoidal amplitude of the phase curve). y_params can also be formatted as a list to make many different plots. A “cleaned” version of y_params will be used in the filenames of the figures and save files relevant for that y_param (e.g. ‘1/r1’ would not work in a filename, so it becomes ‘1-r1’).
y_labels
The formatted string you want on the label of the y-axis. Set to None to use the default formatting which has been nicely formatted in LaTeX for most y_params. If y_params is a list, then y_labels must also be a list unless you want the same value applied to all y_params.
y_label_units
The formatted string for the units you want on the label of the y-axis. For example ‘(ppm)’, ‘(seconds)’, or ‘(days$^{-1})$’. Set to None to automatically add any implied units from y_scalars (e.g. ppm if y_scalars=1e6), or set to ‘’ to force no units. If y_params is a list, then y_label_units must also be a list unless you want the same value applied to all y_params.
y_scalars
This parameter can be used to rescale the y-axis. If set to 100, the y-axis will be in units of percent. If set to 1e6, the y-axis will be in units of ppm. If set to any other value other than 1, 100, 1e6, then the y-axis will simply be multiplied by that value and the scalar will be noted in the y-axis label. If y_params is a list, then y_scalars must also be a list unless you want the same value applied to all y_params.
x_unit
The x-unit to use in the plot. This can be any unit included in astropy.units.spectral (e.g. um, nm, Hz, etc.) but cannot include wavenumber units.
ncol
The number of columns you want in your LaTeX formatted tables. Defaults to 4.
star_Rad
The stellar radius. Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
planet_Teq
The planet’s zero-albedo, complete redistribution equlibrium temperature in Kelvin. Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
planet_Mass
The planet’s mass in units of Jupiter masses. Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
planet_Rad
The planet’s radius in units of Jupiter radii. Set to None to use the average fitted radius. Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
planet_mu
The mean molecular mass of the atmosphere (in atomic mass units). Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
planet_R0
The reference radius (in Jupiter radii) for the scale height measurement. Set to None to use the mean fitted radius. Used to compute the scale height if y_unit is transmission type and isplots_S6>=3.
isplots_S6
Sets how many plots should be saved when running Stage 6. A full description of these outputs is available here: Stage 6 Output.
hide_plots
If True, plots will automatically be closed rather than popping up on the screen.
topdir + inputdir
The path to the directory containing the Stage 5 JWST data. Directories containing spaces should be enclosed in quotation marks.
topdir + outputdir
The path to the directory in which to output the Stage 6 JWST data and plots. Directories containing spaces should be enclosed in quotation marks.
topdir + model_spectrum
The path to a model spectrum to plot underneath the observations to show how the fitted results compare to the input model for simulated observations or how the fitted results compare to a retrieved model for real observations. Set to None if no model should be plotted. The file should have column 1 as the wavelength and column 2 should contain the transmission or emission spectrum. Any headers must be preceded by a #. Directories containing spaces should be enclosed in quotation marks.
model_x_unit
The x-unit of the model. This can be any unit included in astropy.units.spectral (e.g. um, nm, Hz, etc.) but cannot include wavenumber units.
model_y_param
The y-unit of the model. Follow the same format as y_params. If desired, can be rp if y_params is rp^2, or vice-versa. Only one model model_y_param can be provided, but if y_params is a list then the code will only use model_y_param on the relevant plots (e.g. if model_y_param=rp, then the model would only be shown where y_params is rp or rp^2).
model_y_scalar
Indicate whether model y-values have already been scaled (e.g. write 1e6 if model_spectrum is already in ppm).
model_zorder
The zorder of the model on the plot (0 for beneath the data, 1 for above the data).
model_delimiter
Delimiter between columns. Typical options: None (for whitespace), ‘,’ for comma.